고정 헤더 영역
상세 컨텐츠
본문
1. 단항 논리 회귀 실습
- 논리 회귀(Logistic Regrssion)
- 분류를 할 때 사용하며 선형 회귀 공식으로부터 나왔기 때문에 논리 회귀라는 이름이 붙여짐

> 직선 하나(선형 회귀)를 사용해서 예측한다면 제대로 예측할 수 없음

> Sigmoid 함수(Logistic 함수)를 사용하여 정확도를 높임
Sigmoid 함수
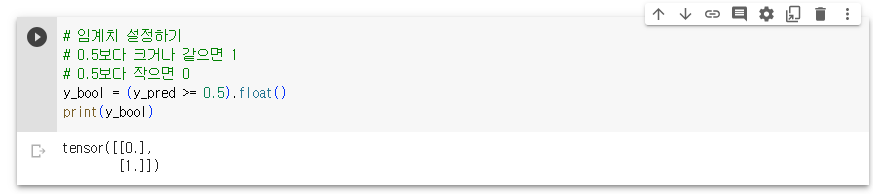
- 예측값을 0에서 1사이 값이 되도록 만듬
- 0에서 1사이의 연속된 값을 출력으로 하기 때문에 보통 0.5를 기준으로 구분







비용 함수
- 논리 회귀에서는 nn.BCELoss() 함수를 사용하여 Loss를 계산
- Binary Cross Entropy




epochs = 1000
for epoch in range(epochs + 1):
y_pred = model(x_train)
loss = nn.BCELoss()(y_pred, y_train)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if epoch % 100 == 0:
print(f'Epoch {epoch}/{epoch} Loss: {loss:.6f}')
2. 다항 논리회귀 실습






3. 와인 품종 예측해보기
sklearn.datasets.load_wine 데이터셋은 이탈리아의 같은 지역에서 재배된 세가지 다른 품종으로 만든 와인을 화학적으로 분석한 결과
문제
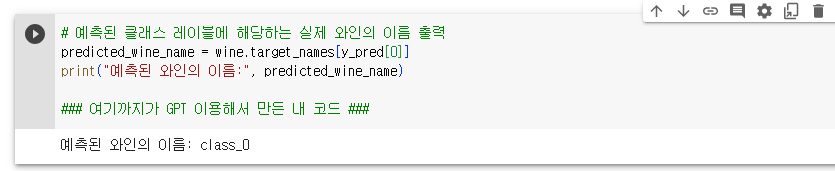
- 13개의 성분을 분석하여 어떤 와인인지 맞춰보자
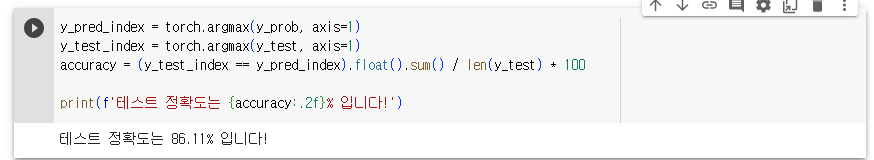
- 단, 트레이닝 데이터를 80%, 테스트 데이터를 20%로 하며 테스트 데이터의 0번 인덱스가 어떤 와인인지 출력하고, 테스트 정확도도 출력
# https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_wine.html#sklearn.datasets.load_wine 에서 갖고온 그냥 예시 코드임
# Author: Virgile Fritsch <virgile.fritsch@inria.fr>
# License: BSD 3 clause
import numpy as np
from sklearn.covariance import EllipticEnvelope
from sklearn.svm import OneClassSVM
import matplotlib.pyplot as plt
import matplotlib.font_manager
from sklearn.datasets import load_wine
# Define "classifiers" to be used
classifiers = {
"Empirical Covariance": EllipticEnvelope(support_fraction=1.0, contamination=0.25),
"Robust Covariance (Minimum Covariance Determinant)": EllipticEnvelope(
contamination=0.25
),
"OCSVM": OneClassSVM(nu=0.25, gamma=0.35),
}
colors = ["m", "g", "b"]
legend1 = {}
legend2 = {}
# Get data
X1 = load_wine()["data"][:, [1, 2]] # two clusters
# Learn a frontier for outlier detection with several classifiers
xx1, yy1 = np.meshgrid(np.linspace(0, 6, 500), np.linspace(1, 4.5, 500))
for i, (clf_name, clf) in enumerate(classifiers.items()):
plt.figure(1)
clf.fit(X1)
Z1 = clf.decision_function(np.c_[xx1.ravel(), yy1.ravel()])
Z1 = Z1.reshape(xx1.shape)
legend1[clf_name] = plt.contour(
xx1, yy1, Z1, levels=[0], linewidths=2, colors=colors[i]
)
legend1_values_list = list(legend1.values())
legend1_keys_list = list(legend1.keys())
# Plot the results (= shape of the data points cloud)
plt.figure(1) # two clusters
plt.title("Outlier detection on a real data set (wine recognition)")
plt.scatter(X1[:, 0], X1[:, 1], color="black")
bbox_args = dict(boxstyle="round", fc="0.8")
arrow_args = dict(arrowstyle="->")
plt.annotate(
"outlying points",
xy=(4, 2),
xycoords="data",
textcoords="data",
xytext=(3, 1.25),
bbox=bbox_args,
arrowprops=arrow_args,
)
plt.xlim((xx1.min(), xx1.max()))
plt.ylim((yy1.min(), yy1.max()))
plt.legend(
(
legend1_values_list[0].collections[0],
legend1_values_list[1].collections[0],
legend1_values_list[2].collections[0],
),
(legend1_keys_list[0], legend1_keys_list[1], legend1_keys_list[2]),
loc="upper center",
prop=matplotlib.font_manager.FontProperties(size=11),
)
plt.ylabel("ash")
plt.xlabel("malic_acid")
plt.show()
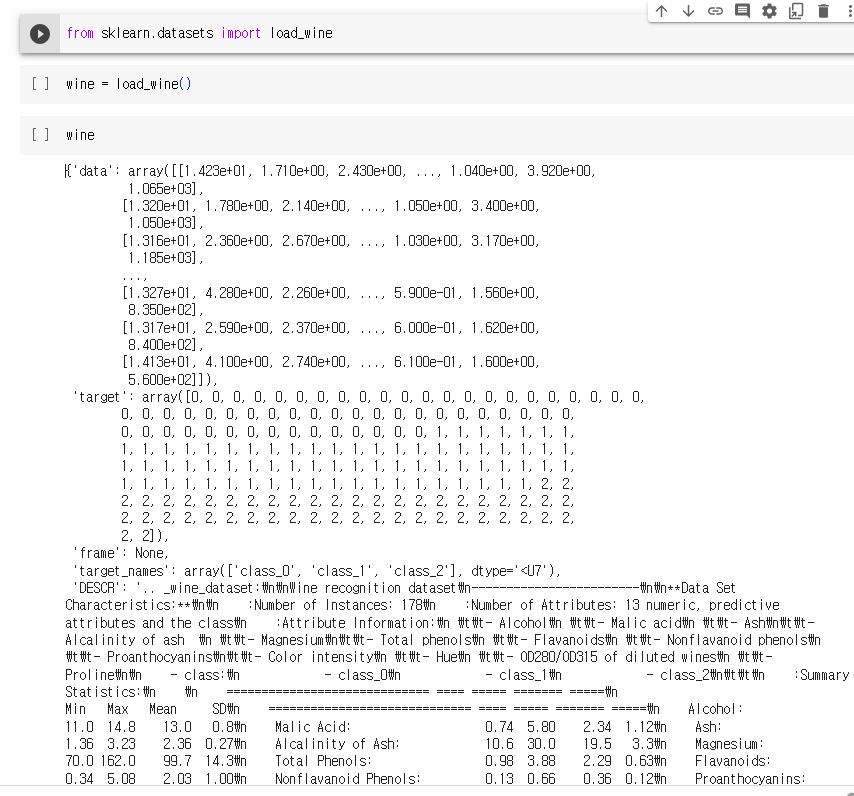
print(wine['DESCR'])








model = nn.Sequential(
nn.Linear(13, 3)
)
optimizer = optim.Adam(model.parameters(), lr=0.01)
epochs = 1000
for epoch in range(epochs + 1):
y_pred = model(x_train)
loss = nn.CrossEntropyLoss()(y_pred, y_train)
optimizer.zero_grad()
loss.backward()
optimizer.step()
if epoch % 100 == 0:
y_prob = nn.Softmax(1)(y_pred)
y_pred_index = torch.argmax(y_prob, axis=1)
y_train_index = torch.argmax(y_train, axis=1)
accuracy = (y_train_index == y_pred_index).float().sum() / len(y_train) * 100
print(f'Epoch { epoch:4d}/{epochs} Loss: {loss:.6f} Accuracy: {accuracy:.2f}%')
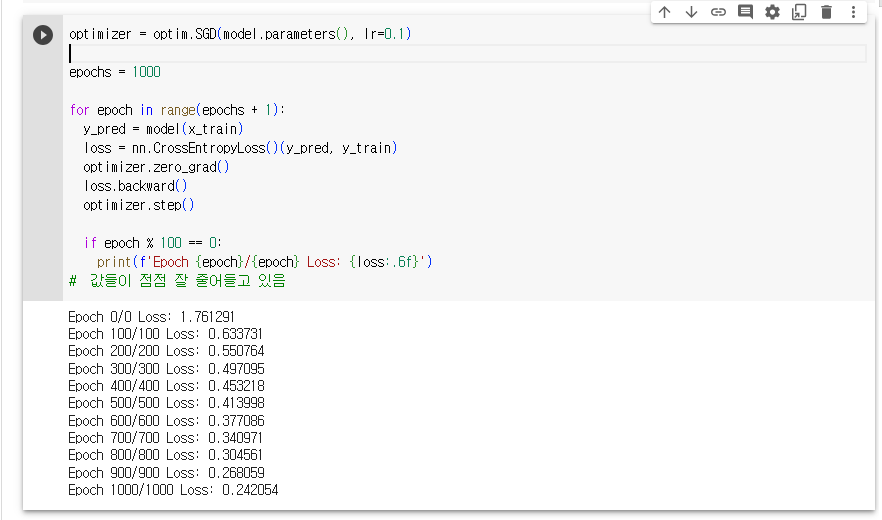
# 정확도가 낮은걸 보니, 경사하강법을 사용하자.
# SGD 에서 Adam으로 바꾸니 정확도가 확 올라감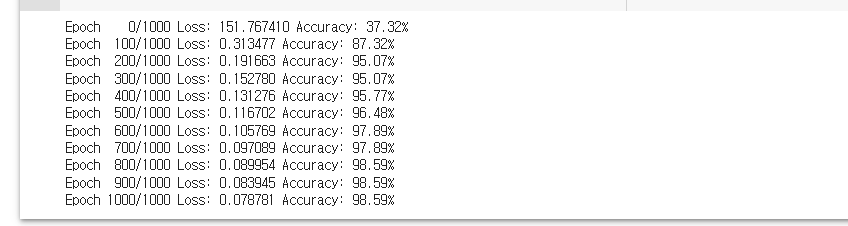
4. 경사하강법의 종류
4-1. 배치 경사하강법
- 가장 기본적인 경사하강법(Vanilla Gradient Descent)
- 데이터셋 전체를 고려하여 손실함수를 계산
- 한 번의 Epoch에 모든 파라미터 업데이트를 단 한번만 수행
- Batch의 개수와 Iteration은 1이고, Batch size는 전체 데이터의 개수
- 파라미터 업데이트할 때 한 번에 전체 데이터셋을 고려하기 때문에 모델 학습시 많은 시간과 메모리가 필요하다는 단점
4-2. 확률적 경사하강법
- 확률적 경사하강법은 배치 경사하강법(Stochastic Gradient Descent)이 모델 학습시 많은 시간과 메모리가 필요하다는 단점을 계산하기 위해 제안된 기법
- Batch size를 1로 설정하여 파라미터를 업데이트 하기 때문에 배치 경사하강법 보다 훨씬 빠르고 적은 메모리로 학습이 진행
- 파라미터 값의 업데이트 폭이 불안정하기 때문에 정확도가 낮은 경우가 생길 수 있음
- SGD가 여기에 포함됨
4-3. 미니 배치 경사하강법
- 미니 배치 경사하강법(Mini-Batch Gradient Descent)은 Batch size가 1도 전체 데이터 개수도 아닌 경우
- 배치 경사하강법 보다 모델 학습 속도가 빠르고, 확률적 경사하강법 보다 안정적인 장점이 있음
- 딥러닝 분야에서 가장 많이 활용되는 경사하강법
- 일반적으로 Batch size를 32, 64, 128과 같이 2의 n제곱에 해당하는 값으로 사용하는게 보편적
5. 경사하강법의 여러가지 기술들
5-1. 확률적 경사하강법(SGD)
- 매개변수 값을 조정 시, 전체 데이터가 아니라 랜덤으로 선택한 하나의 데이터에 대해서만 계산하는 방법
5-2. 모멘텀(Momentum)
- 관성이라는 물리학의 법칙을 응용한 방법
- 경사하강법의 관성을 더해줌
- 접선의 기울기에 한 시점 이전의 접선의 기울기 값을 일정한 비율만큼 반영
- 언덕에서 공이 내려올 때 중간의 작은 웅덩이에 빠지더라도 관성의 힘으로 넘어서는 효과를 줄 수 있음
5-3. 아다그라드(Adagrad)
- 모든 매개변수에 동일한 학습률(Learning rate)을 적용하는 것은 비효율적이라는 생각에서 만들어진 학습 방법
- 처음에는 크게 학습하다가 조금씩 작게 학습시킴
5-4. 아담(Adam)
- 모멘텀 + 아다그라드